Meta-Analysis Specification
Now that your studyset's studies have all the necessary information (metadata, coordinates, annotations), you can configure and run a meta-analysis!
A wizard will guide you through the following key steps of meta-analysis specification.
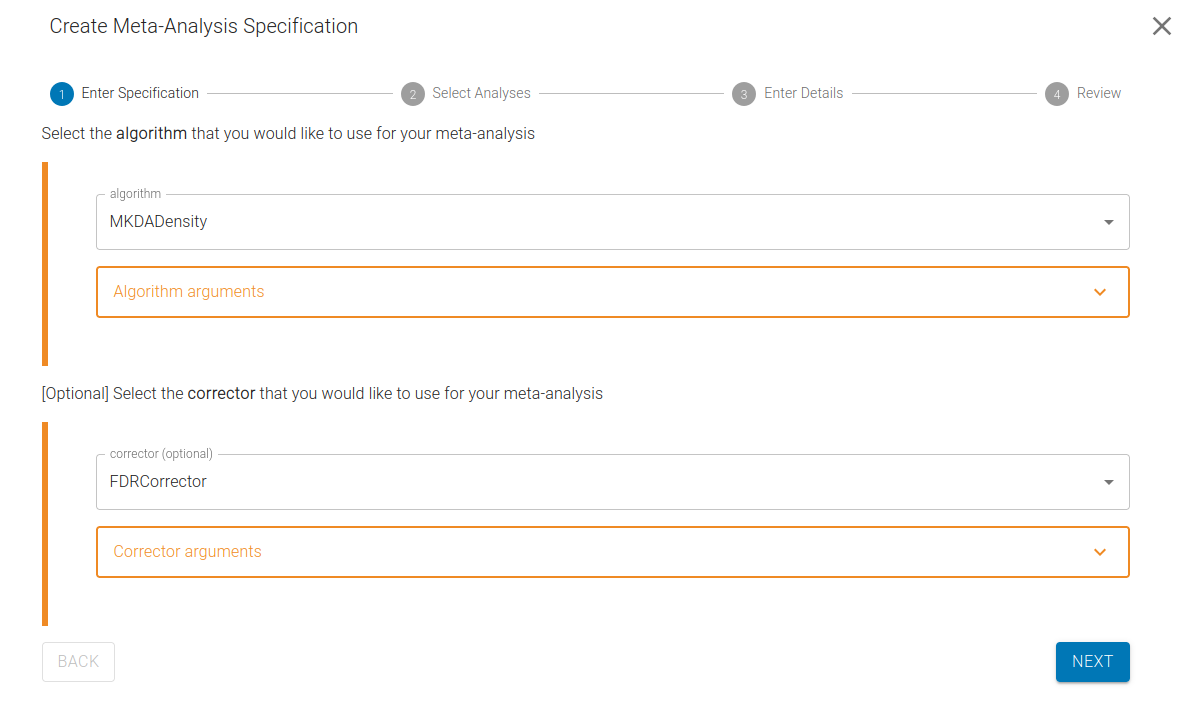
1) Select the Algorithm and Corrector
Choose the algorithm for your meta-analysis and, optionally, select a statistical correction method (corrector).
Neurosynth-Compose supports several meta-analysis algorithms, powered by NiMARE, a Python library for neuroimaging meta-analysis. For an overview to the coordinate-based meta-analysis methods supported by NiMARE, refer to this guide.
Supported coordinate-based methods include:
MKDADensity (default): Multi-Kernel Density Analysis. A popular density-based meta-analysis algorithm. Creates a binary sphere around each coordinate.
ALE: Activation Likelihood Estimation, convolves coordinates with a 3D Gaussian distribution.
MKDAChi2: Allows you to compare your meta-analysis studies with a larger reference set of studies. It tests whether a higher proportion of studies in your meta-analysis activate a specific voxel compared to a larger population of studies that were not included. For more details, see the MKDAChi2 Association tutorial
For each algorithm, a default set of arguments is used, but you can easily modify them. For a complete reference on available arguments, see the NiMARE API Docs.
Next, select a method for multiple comparisons correction:
- FDRCorrector (default): False Discovery Rate correction.
- FWECorrector: Family-Wise Error Rate correction. We strongly recommend using FWECorrector for publication-quality results with 10,000s iteration if possible. This is computationally intensive however, hence not the default option.
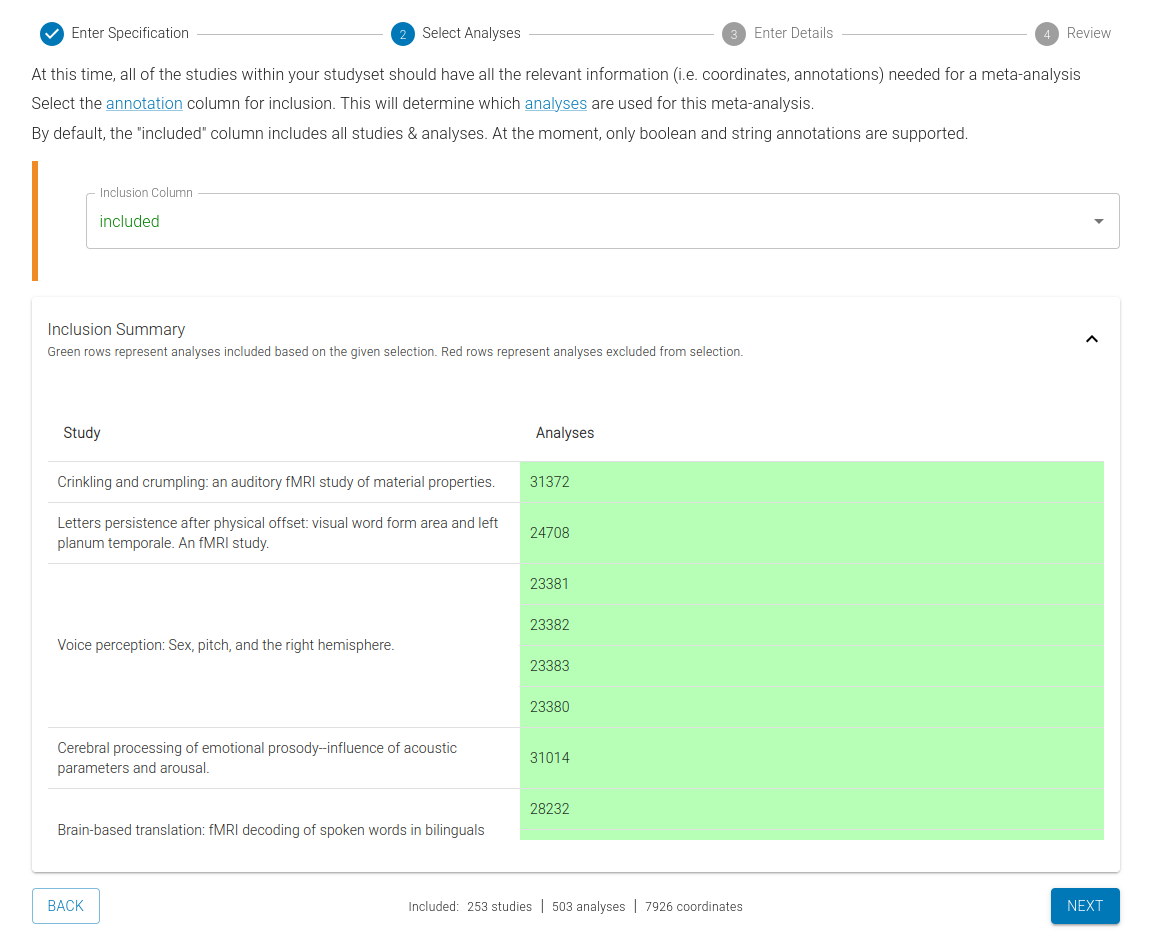
2) Select Analyses
Now you will select which studies & analyses you want to include in your meta-analysis. To do so, select the annotation column for inclusion, which determines which analyses are used in the meta-analysis. By default, the "included" column contains all studies and analyses, but you can modify this based on your needs by editing Annotations in the Extraction step.
A preview of the included studies based on the selected annotation column will be displayed.
3) Meta-Analysis Details
Enter a name and description for your meta-analysis to help you find it later. A default name will be provided, but you can customize it as needed.
4) Review
Finally, you can review your selected options before creating your specification.
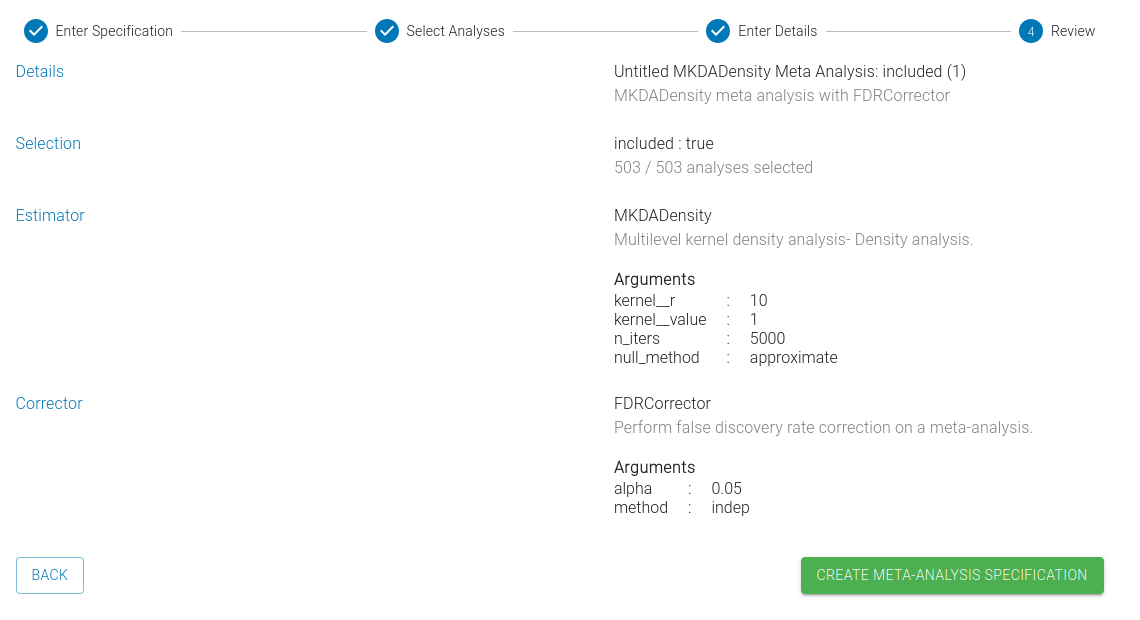
5) Finalize
To complete the specification process, click the "Create Meta-Analysis Specification" button.
Running the Meta-Analysis
After clicking "Create Meta-Analysis Specification," you will be taken to a page displaying the status of your running analyses. Refer to the next section of the documentation for details on running and monitoring your meta-analysis.