Running Analyses
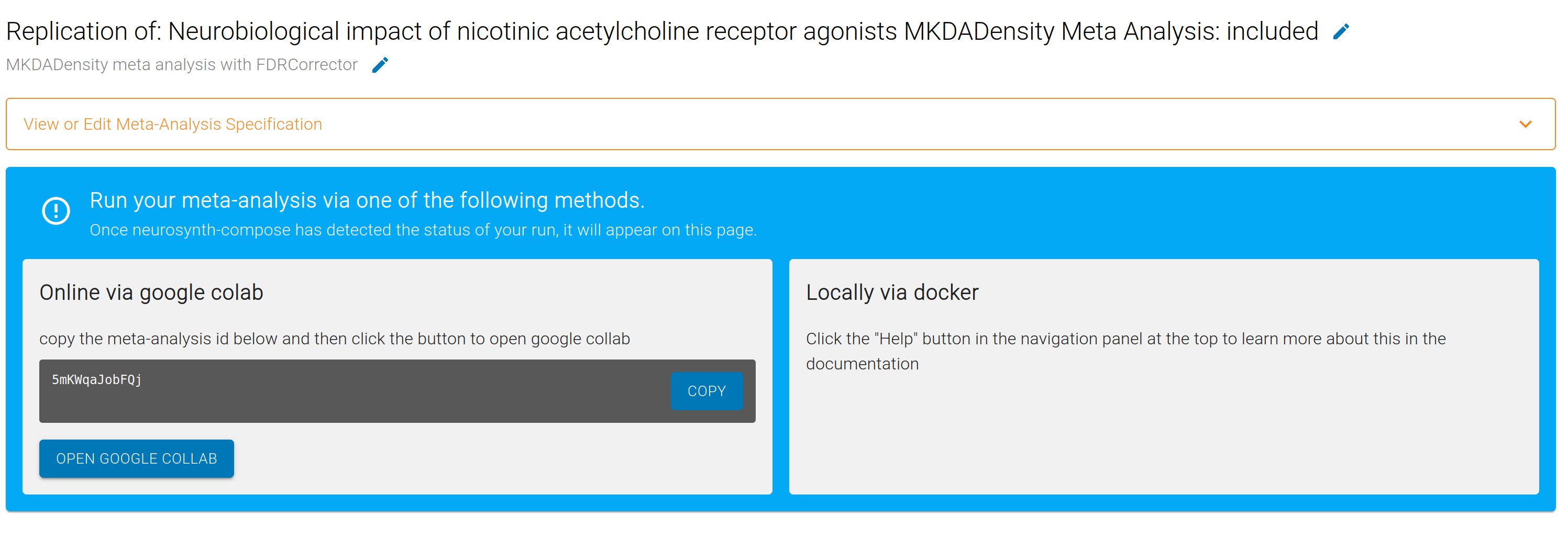
You have a several options for running the analysis. In all cases, you will need your unique <meta-analysis-id>, which you can access for each Meta-Analysis within your Project.
Under the hood, analyses are managed by the nsc-runner Python package, and executed by the NiMARE (Neuroimaging Meta-Analysis Research Environment) Python package.
Google Colab
The easiest way to run an analysis is to use the Google Colab notebook linked above.
The provided notebook runs entirely in the cloud, and does not require any local installation of software.
To use simply paste your analysis ID into the first cell (META_ID), and using the Toolbar selet (Runtime -> Run All)
or the keyboard shortcut (Ctrl or ⌘ + F9) to run the notebook.
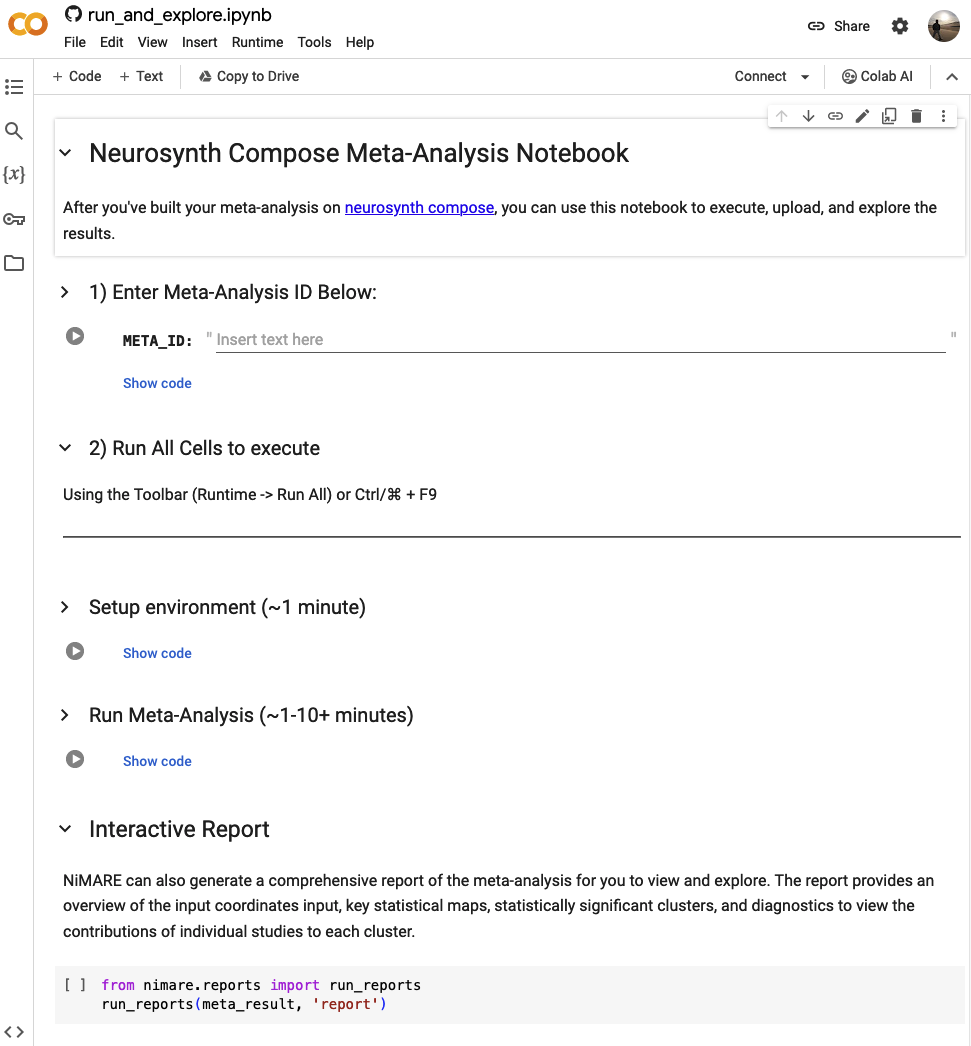
The notebook will install all required software, run the analysis, and upload the results to Neurosynth Compose. Once the analysis is complete, you can use the notebook to explore the results using the interative report, download an archive of the results, or browse the results in the Neurosynth Compose web interface, in the Meta-Analysis section of your Project.
The Colab notebook has limited and varying freely available resources, and may not be able to run large analyses. If your analysis fails, try running it again, or using one of the other methods below.
Docker
The easiest way to run analyses locally is to use the nsc-runner Docker image provided by Neurosynth Compose.
Docker is a containerization technology that allows you to run software in a consistent environment, regardless of the underlying operating system.
To run the Docker image, you will need to install Docker on your local machine. Instructions for installing Docker can be found here.
Once Docker is installed, you can run your analysis using the using the following command:
docker run -it -v -v /local/dir:/results ghcr.io/neurostuff/nsc-runner:latest <meta-analysis-id>
where /local/dir is the path to a local directory where you would like to save the results of your analysis, and <meta-analysis-id> is the ID of the meta-analysis you would like to run.
If you are trying to run the analysis from a mac with an ARM chip (eg an M1 Macbook air) use this command instead:
docker run -it --platform=linux/amd64 -v /local/dir:/results ghcr.io/neurostuff/nsc-runner:latest <meta-analysis-id>
The Docker image will download all required software, run the analysis, and upload the results to Neurovault & Neurosynth Compose. An HTML report will be saved in the results directory, and the results will be available in the Meta-Analysis section of your Project on Neurosynth Compose.
Updating the Docker image
For every release of nsc-runner, we publish a corresponding Docker image.
You can manually download a specific neuroscout-cli release as follows:
docker pull ghcr.io/neurostuff/nsc-runner:<version>
where <version> is the version of nsc-runner that you want to download. If you omit version, the latest stable version will be downloaded.
You can see the tags available for download on GitHub
Manually prepared environment using pip
Manually installing nsc-runner may be difficult due to complex dependencies in the SciPy stack, or fMRI-specific tooling. Proceed only if you know what you’re doing.
Use pip to install nsc-runner from PyPI:
pip install nsc-runner
and then run the analysis using the following command:
nsc-runner <meta-analysis-id>
Viewing and intepreting your results
After successfully running your meta-analysis, results will be uploaded back to Neurosynth Compose and NeuroVault, where you can interactively view and interpret your results. See the next section in the documentation for more detais.